Pharos-3D
Overview
Example Result
Getting Started
Access and Pricing
References
Overview
Pharos-3D is Alipheron’s technology to screen ultra-large combinatorial spaces for molecules with a similar shape and similar pharmacophore profile to a given query structure. Typically, this query structure is a bio-active conformation of a small molecule known to inhibit a specific target protein. Pharos-3D identifies those molecules of a combinatorial space that possess at least one low-energy conformer that matches the query conformer’s shape nicely and at the same time may interact with a target protein in a similar way due to similar locations of pharmacophoric features.
Pharos-3D searches are computationally demanding, because they involve the generation of conformers of thousands of partially assembled and also completely enumerated structures. A typical result of a Pharos-3D search contains thousands of molecules with 3-dimensional atom coordinates that match well the query structure. Typically, these molecules are ranked then by another method, e.g. ligand-protein docking, to determine a most promising subset to be ordered for synthesis at the provider of the space.
Pharos-3D docked by DataWarrior into binding site of PDB-entry ‘4FFW’
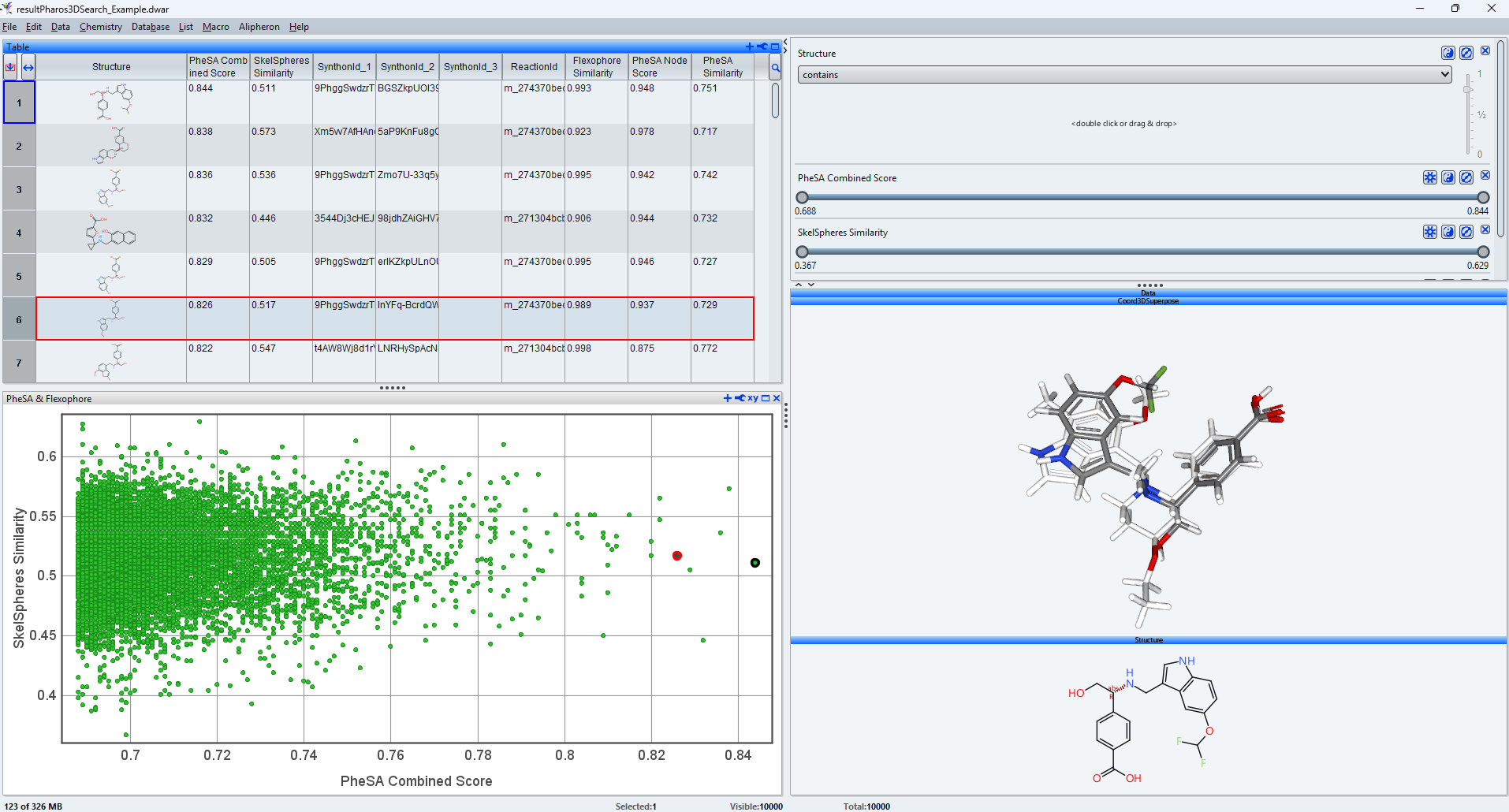
Example Result
The PheSA Combined Score is the key metric to assess fit to the query compound. The score ranges from 0 to 1, indicating “zero” and “identical shape and pharmacophoric features”, respectively.
The SkelSphere Similarity metric is a powerful measure describing structure similarity from a chemist’s perspective. It also ranges from 0 to 1, indicating “zero similarity” and “identity”, respectively.
Getting Started
Running a Pharos-3D search is not much different than a Hyperspace search. Make sure, you have installed DataWarrior version v06.04.01 or newer. Then, install the Pharos-3D plugin, either by selecting Alipheron Pharos-3D from the Help->Trusted Plugins menu, or, if you have received a dedicated plugin for your own space, by putting that plugin-jar file into the DataWarrior plugin folder. Then, quit DataWarrior and re-launch it to get a new Alipheron->Pharos-3D Search… menu item. Select this item and the following dialog opens.
After selecting the space to be searched, you need to define your query conformer. A right mouse click in the query field opens a menu letting you paste in or load a molecule from a file or a ligand directly from the PDB database. You may change the protonation state of acidic or basic atoms and assign lower or higher importance to parts of the query conformer. Pressing OK submits your query definition to the server, which immediately starts to work on you request.
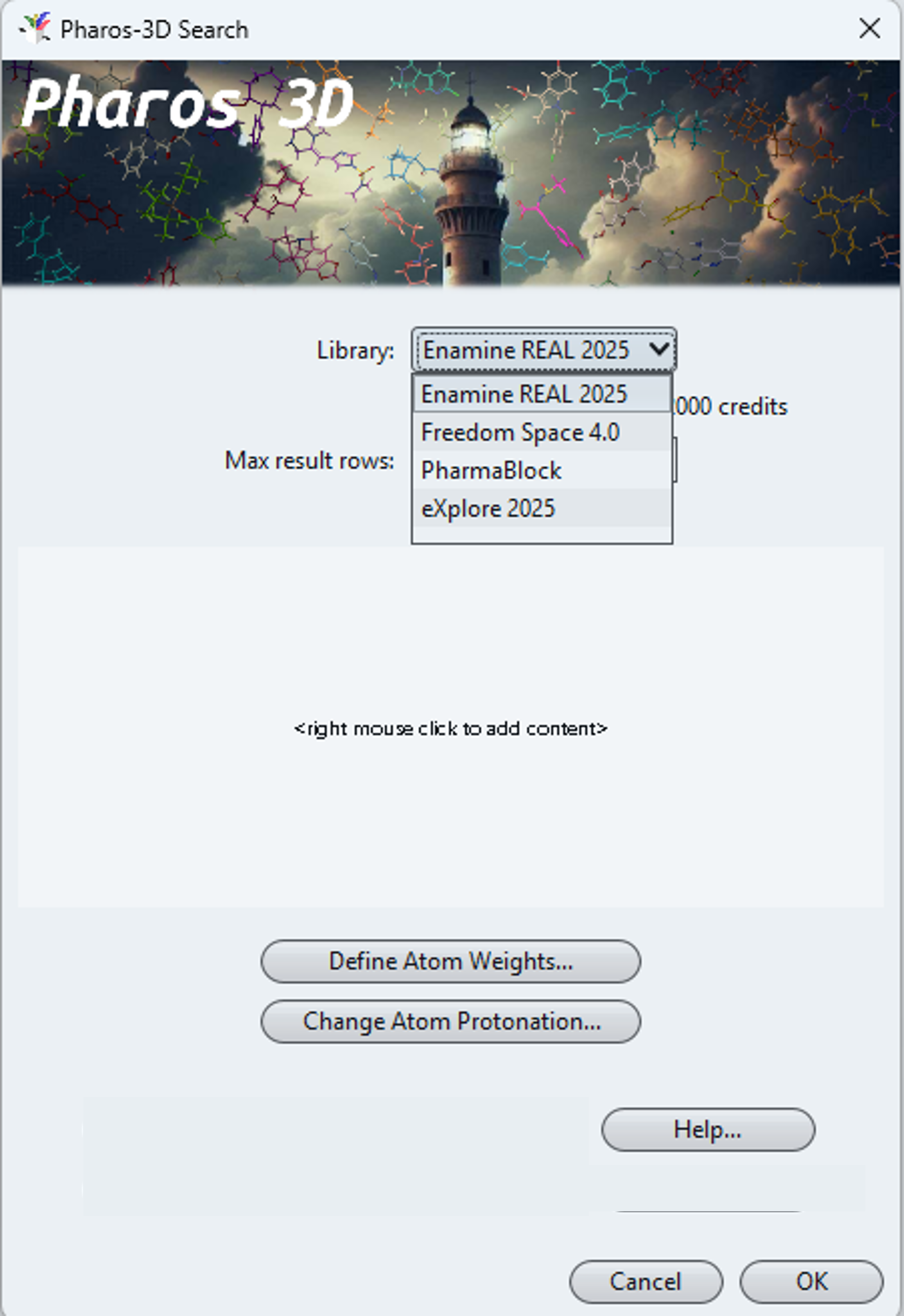 Easily usable plugin from within DataWarrior. Input is a 3-dimensional molecular structure, ideally a bioactive conformer.
Easily usable plugin from within DataWarrior. Input is a 3-dimensional molecular structure, ideally a bioactive conformer.
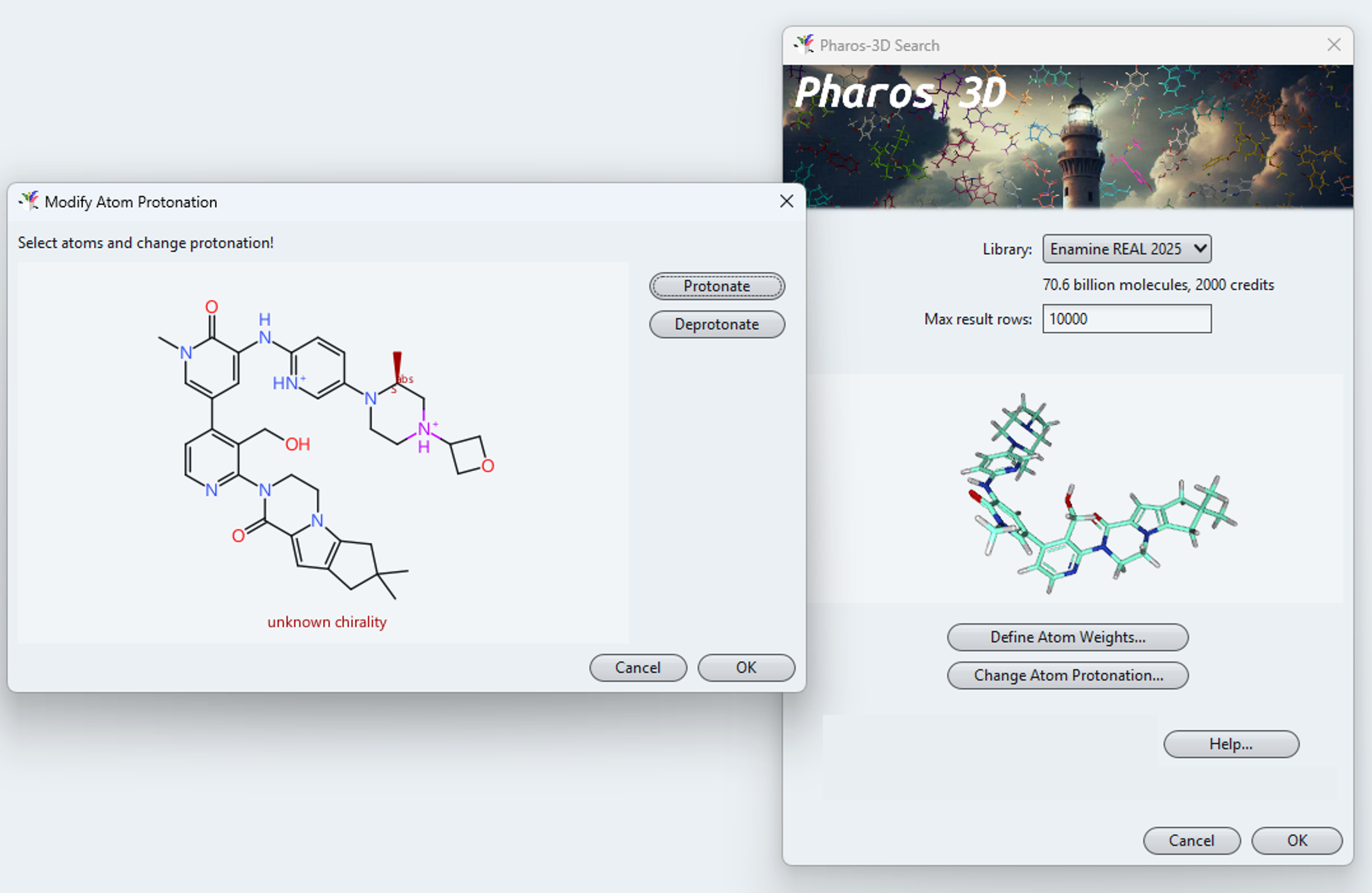 Protonation states can be configured.
Protonation states can be configured.
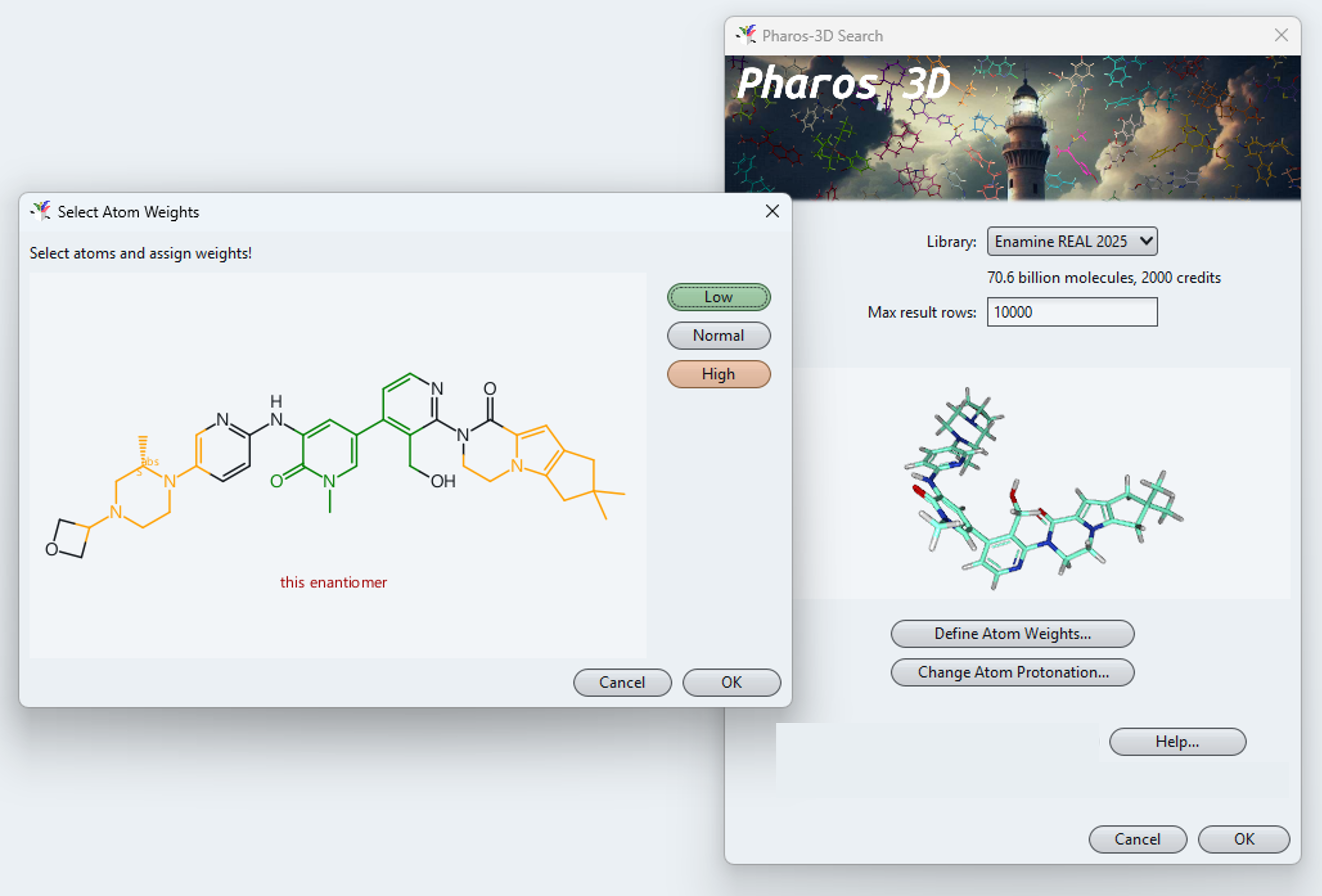 Partial structural weighting can be applied to fine-tune the shape alignment.
Partial structural weighting can be applied to fine-tune the shape alignment.
Typically, three to ten hours later you receive an e-mail with a link to download
the search result as a DataWarrior file. A successful search result contains up to 10'000
result molecules, which are all aligned in space to the query molecule to allow a quick judgement
about their 3D-similarity to the query. Typically, the next step is then to use your own
criteria for ranking those results to order a most promising subset for synthesis.
If the target structure is known, then a protein-ligand docking may be performed to determine
the most promising subset. This can be done within DataWarrior or with your favorite docking tool.
Other citeria as molecular flexibility, lipophilicity, solubility, bio-availability, structure
diversity, and ligand based screening methods may also be considered to rank these compounds.
Access and Pricing
For access to Pharos-3D, please register with your name and e-mail address. Searches with Pharos-3D require either purchased tokens or a licensing agreement. Tokens can be purchased from Paypal or specifying a credit card. Pharos-3D can be installed as an in-house solution that offers private searches on the Alipheron ULCL Collection. In addition, this option enables searches on your company’s custom-built in-house spaces, which may be built from your selection of reactions and your proprietary building blocks. For licensing options, reach out to us at contact@alipheron.com.
References
-
Flexophore, a new versatile 3D pharmacophore descriptor that considers molecular flexibility; M von Korff, J Freyss, T Sander; Journal of Chemical Information and Modeling, 2008, 48 (4), 797-810; https://doi.org/10.1021/ci700359j
-
PheSA: An Open-Source Tool for Pharmacophore-Enhanced Shape Alignment; J Wahl; Journal of Chemical Information and Modeling, 2024, 64 (15), 5944-5953; https://doi.org/10.1021/acs.jcim.4c00516